Members of Phosphodiesterase (PDE) Superfamily Hydrolyze Cyclic Nucleotides
Three major classes of cyclic nucleotide phosphodiesterases (PDEs) have been discovered, each with its own distinctive catalytic domain structure:
- Class I PDEs contain the characteristic PDEase I catalytic domain and consist of eleven distinct families differing in substrate specificity, tissue localization, regulatory mechanisms, and pharmacological properties.
- Class II PDEs have been identified in slime molds, fungi, and bacteria, and contain a distinct catalytic domain structure (PDEase II) that is selective for cAMP as a substrate.
- Class III PDEs exist only in prokaryotes, and have a completely different catalytic mechanism that resembles that of the purple acid phosphatases.
cAMP and cGMP Hydrolyzing PDEs in Vertebrates
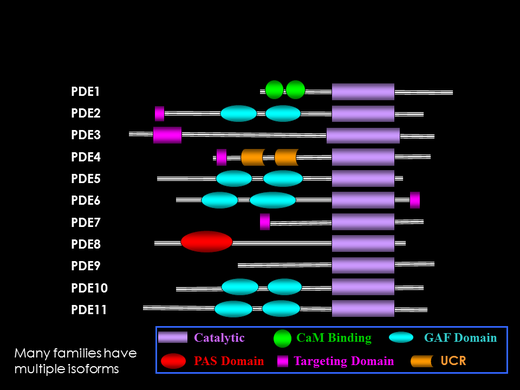
Eleven families of Class I PDEs have been identified in vertebrates. Some families are multi-genic; for example, the PDE6 family consists of 3 genes, PDE6A (rod), PDE6B (rod), and PDE6C (cone).
All eleven PDE families contain a homologous C-terminal domain (purple box in figure) that represents the catalytic pocket of the enzyme. Differences in the amino acid sequence of the different PDE families are known to account for the substrate specificity (cAMP-specific, cGMP-specific, or dual-substrate) and pharmacological properties that distinguish the different enzyme families.
The N-terminal, regulatory domain of PDE is what best distinguishes the eleven PDE families. Although not yet demonstrated for each PDE family, it is generally assumed that changes in the N-terminal region of PDE induced by post-translational modifications or ligand binding result in changes in the catalytic domain, thus modulating enzyme activity.
Five of the eleven PDE families (including the photoreceptor PDE6 family) contain regulatory modules that have been termed GAF domains (turquoise ovals in figure). Several of the GAF-containing PDEs bind cGMP at noncatalytic binding sites, resulting in allosteric regulation of enzyme activity in the catalytic domain. Our lab studies the molecular evolution of PDEs to better understand structure, function, and regulation of this enzyme superfamily.
All eleven PDE families contain a homologous C-terminal domain (purple box in figure) that represents the catalytic pocket of the enzyme. Differences in the amino acid sequence of the different PDE families are known to account for the substrate specificity (cAMP-specific, cGMP-specific, or dual-substrate) and pharmacological properties that distinguish the different enzyme families.
The N-terminal, regulatory domain of PDE is what best distinguishes the eleven PDE families. Although not yet demonstrated for each PDE family, it is generally assumed that changes in the N-terminal region of PDE induced by post-translational modifications or ligand binding result in changes in the catalytic domain, thus modulating enzyme activity.
Five of the eleven PDE families (including the photoreceptor PDE6 family) contain regulatory modules that have been termed GAF domains (turquoise ovals in figure). Several of the GAF-containing PDEs bind cGMP at noncatalytic binding sites, resulting in allosteric regulation of enzyme activity in the catalytic domain. Our lab studies the molecular evolution of PDEs to better understand structure, function, and regulation of this enzyme superfamily.
